I first started looking for how to implement a model II nested random effects ANOVA (as described in Bailey and Byrnes 1990), like SAS's PROC NESTED but in R, and ran into the usual problems trying to get the two programs to give equivalent output (where were my mean squares in R? So aggravating sometimes...). I was also looking to see exactly how a similar concept that I just learned about, repeatability (Lessells and Boag 1987; also known as the intraclass correlation coefficient), was related. At first I thought they were the same thing, but after carefully writing out the equations from both Bailey and Byrnes (1990) and Lessells and Boag (1987), I realized they can be calculated easily from one another (and I was backed up by further reading in Wolak et al. 2012, pg. 130, who state "measurement error is simply one minus the repeatability", as I had suspected). Measurement error uses within-groups mean square error (or variance within groups) as the numerator in the calculation, while the intraclass correlation coefficient (or repeatability) uses the among-group variance (which both Lessells and Boag 1987 and Bailey and Byrnes 1990 show to calculate using the same method).
As an aside, repeatability appears to be used rather broadly, and intraclass correlation coefficient also appears to have some other defintions, if wikipedia's summary is to be believed, but in the papers I am citing they all are comparing within and among group variation (the ANOVA framework).
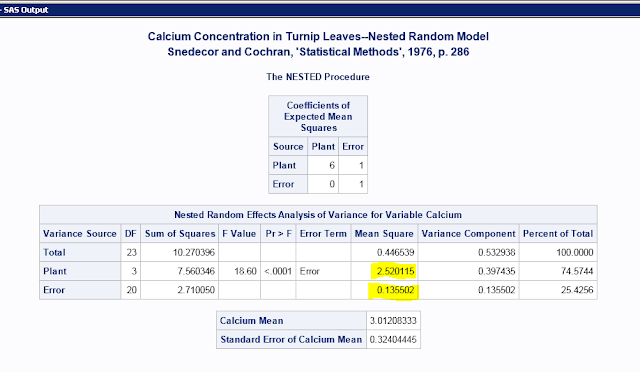
Armed with this new terminology and knowledge, I decided to use the R package 'ICC' (Wolak et al. 2012) which calculates repeatability. I tested this on some sample data from the SAS website with both the model II nested ANOVA as used in Bailey and Byrnes (1990) and in R.
Use the "turnip" dataset as the SAS website shows, then run this modified code:
proc nested data=turnip;
classes plant;
var calcium;
run;
To get this:
Calculate repeatability with these highlighted numbers using is the equation from Lessells and Boag (1987); 6 is the number of samples per individual, i.e. the number of repeated measurements:
((2.520115-0.135502)/6)/(0.135502+((2.520115-0.135502)/6))
Repeatability=0.745745
To calculate measurement error, substitute 0.135502 for the current numerator (i.e. use the Bailey and Byrnes 1990 equation).
0.135502/(0.135502+((2.520115-0.135502)/6))
Measurement error=0.254255
Adding the two numbers together gets 1, which means all the variation is accounted for (and thus you only need to calculate one; to get the other just subtract from 1).
Using the ICC package's function ICCest() gives essentially the same answer (the difference appears to be the level of precision in the decimal places), but also provides confidence intervals and other data that are explained more in the Wolak et al. paper. (Import the data first, obviously, before running this code. You can enter it by hand or export from SAS as a .csv).
ICCest(x=plant, y=calcium, data=turnip)
>$ICC
>[1] 0.7457443
>
>$LowerCI
>[1] 0.3889918
>
>$UpperCI
>[1] 0.9776527
>
>$N
>[1] 4
>
>$k
>[1] 6
>
>$varw
>[1] 0.1355025
>
>$vara
>[1] 0.3974355
Finally, since SAS uses a random effects model, I hoped that one might be able to calculate measurement error by hand from R just to be sure it all works. I found a page comparing different methods of repeatability in R that showed an example using the package 'lme4'. Calcium is the measured variable, plant is the random effect, and there are no fixed effects.
model.repeat<-lmer(calcium~1+(1|plant),
data=turnip)
summary(model.repeat)
>results>Linear mixed model fit by REML t-tests use Satterthwaite >approximations to degrees
> of freedom [lmerMod]
>Formula: calcium ~ 1 + (1 | plant)
> Data: turnip
>
>REML criterion at convergence: 31.2
>
>Scaled residuals:
> Min 1Q Median 3Q Max
>-1.09999 -0.85753 -0.09009 0.69564 2.13276
>
>Random effects:
> Groups Name Variance Std.Dev.
> plant (Intercept) 0.3974 0.6304
> Residual 0.1355 0.3681
>Number of obs: 24, groups: Plant, 4
>
>Fixed effects:
> Estimate Std. Error df t value Pr(>|t|)
>(Intercept) 3.012 0.324 3.000 9.295 0.00264 **
>---
>Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Use the residual (error, or within) as the numerator if you want to calculate measurement error, or the plant (among groups) variance as the numerator if you want to calculate repeatability. For example, for measurement error, you get pretty close to the SAS-by-hand number.
0.1355/(0.3974+0.1355)
Measurement error=0.2542691
So, all three methods (by hand with lme4 in R, using the ICC package in R, and by hand from SAS PROC NESTED) give comparable results for the balanced design (for the plant dataset, n=6 for every individual plant, i.e. there are six measurements for any given plant). For an unbalanced design, with varying numbers of measurements per group, Lessells and Boag (1987) provide an equation to calculate the appropriate sample size coefficient n0 (see their equation 5). The ICC package uses that equation to calculate n0 (Wolak et al. 2012; this is listed as $k in the output from ICCest) I tested this in the R packages only as I was hoping the variance from lme4 might have already been adjusted for the sample size coefficient issue. ICC already states that they adjust (Wolak et al. 2012 and in the help file). With the first row deleted from the turnip data set, ICC gives a repeatability value of 0.7432083 and k (which is n0 from Lessells and Boag)= 5.73913 (slightly under the previous 6, which makes sense as I deleted one row). With that same one-row-deleted data using lme4, I took the random effects variances (0.3952/(0.1419+0.3952)) and got 0.7358034, so its variance calculation is not including the repeatability adjustment for n0. This is not really surprising since that's not what it's designed to do.
So, what I will be doing to check for "measurement error" on my birds (how much "wobble" each bird has in its otherwise standard songs) is substitute the song variable (let's say frequency) for "calcium" and the bird ID (individual ID or band number) for "plant". The random effect in lmer, the classes in SAS PROC NESTED, and x in ICCest are all the individual/subject/item from which you get repeated measurements for measurement error or repeatability.
Take home message: ICC is the easiest to use as it will take into account an unbalanced design for you already. With SAS you can calculate for an unbalanced design but you'll have to do it by hand. Thus, ICC and SAS should cover all cases. With lme4 you can calculate repeatability and measurement error for balanced designs by hand, but I don't see a particularly obvious way to get the mean squares as in SAS (and I need to move on to other parts for the analysis), so I'm going to leave it at that for now.
Here are the papers I used to understand how to calculate measurement error and repeatability:
Bailey RC, Byrnes J. 1990. A new, old method for assessing measurement error in both univariate and multivariate morphometric studies. Systematic Biology 39:124–130.
Wolak ME, Fairbairn DJ, Paulsen YR. 2012. Guidelines for estimating repeatability. Methods in Ecology and Evolution 3:129-137.
and here are some helpful examples of papers that use measurement error:
Lougheed SC, Arnold TW, Bailey RC. 1991. Measurement error of external and skeletal variables in birds and its effect on principal components. Auk 108:432–436.
Marantz CA, Patten MA. 2010. Quantifying subspecies analysis: a case study of morphometric variation and subspecies in the woodcreeper genus Dendrocolaptes. Ornithological Monographs 67:123–140.
No comments:
Post a Comment
Comments and suggestions welcome.